Research Programme: Epigenetic regulation mediated by p63 in epithelial tumours
Principal Investigator: Prof. Eleonora Candi

Scientific interest
Eleonora’s main research interests focused on understanding the molecular mechanisms regulating epithelial development in normal and pathological conditions. She used different approaches from protein chemistry/enzymology to cell/molecular biology and animal models. Originally, she worked on the molecular mechanisms of corneification, a specialized form of cell death in the skin. She biochemically studied the major players involved in cornified cell envelope assembly, including enzymes (TGase 1,3 and 5) and its substrates (loricrin, SPRs and keratins) and how their mutations cause human genetic skin pathologies (Candi et al. Nat Rev Mol Cell Biol. 2005). Recently, Eleonora’s research interest is focused on the transcription factor p63, member of the p63 family. p63 is a crucial signalling node during epidermal differentiation and stem cells maintenance, while in tumours it can engage specific oncogenic programs related to apoptosis resistance and promotion of anchorage-dependent and-independent growth, mobility and invasion in squamous cell carcinomas (SCC) of different origins (lung, skin, head and neck). Her major contributions included studies on p63 transgenic mice (Candi et al CDD 2006, Candi et al PNAS 2007), identification of miRNAs/lncRNAs controlling p63 in stemness/differentiation/senescence (Lena et al CDD 2006, Rivetti PNAS 2012, Panatta et al EMBO Reports 2020). Currently, using knockout mouse models and mass-spec analysis/biochemistry, she is defining the p63 interactome and how p63-dependent altered epigenetic events deeply influence SCC biology.
Current Research Direction
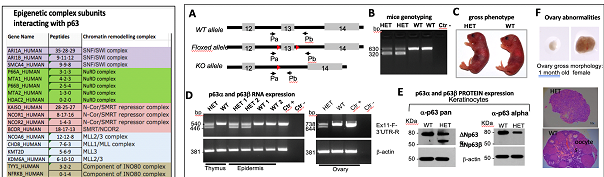
p63-dependent modulation of the chromatin landscape of its target genes is mediated by interaction with specific factors/complexes, therefore defining p63α interactome is of crucial relevance to uncover novel p63α oncogenic role. The p63α isoform is the longest one, containing the protein-protein interacting domain SAM and the TID domain. The majority of the protein-protein interactions engaged by p63α involve its C-terminus, indicating that the p63a C-terminus is the crucial sensor to establish a p63a-selective transcriptional program. To explore the functional relevance of p63α and its interactors we have recently generated a selective p63α knock-out model deleting the SAM domain. The long-term goal of the current proposal is to accomplish a deeper understanding of the p63a interactome and to define p63a-dependent epigenetic regulations in cancer cells. We will study the oncogenic role of the p63 C-terminus, focusing on SAM and TID domains. As experimental model, we will use human SCC cell lines and the new generated p63-SAM knock-out mouse.
To understand the role of p63a as oncogene and to foresee novel therapeutic strategies, is mandatory to:
Task 1: Define p63a interactome in cancer cells. To understand how p63a C-terminus mechanistically recruits chromatin remodeller/epigenetic modulators to its target genes;
Task 2: Define the in vivo function of p63a isoform to unravel the role of the SAM and TID domain. Using the selective p63α-SAM knockout mouse model, we aim to investigate the functional contribution of the p63α isoforms. Particular emphasis will be given to unveil a role for the SAM domain in mediating the oncogenic role of p63 in SCC.
Relevance for cancer
This research fills a gap in cancer epithelial biology, providing an innovative edge for potential therapeutic exploitation. It advances our field, currently lacking robust, high-throughput technologies to determine the spatiotemporal organization of epithelial transcription-translation, affecting cancer cell fate via epigenetics regulation. Insight from new mouse models will advance development of novel technologies-protocols that will enable other researchers to apply our methods to their studies. Results will provide advanced stratification rational for patient’s prognosis.